03 - custom curation database
Build custom databases for protein-coding gene curation
By default, MitoPilot uses protein-coding gene sequences from NCBI RefSeq to finetune start and stop codon positions of your annotations.
We have provided a helper function MitoPilot::custom_curation_db, which allows you to supplement the RefSeq databases with your own protein-coding gene sequences. Using a custom database can greatly improve automatic curation if your focal clade is poorly represented in RefSeq.
You could gather additional protein-coding gene sequences from many sources, including:
- GenBank mitogenomes that are not part of RefSeq
- mitogenomes from other data repositories
- your own unpublished mitogenomes
Make sure to carefully consider what you are adding to the custom database. You should only use high-confidence sequences, as poor quality reference data will result in poorly curated gene models.
Please see the MitoPilot::custom_curation_db documentation for further instructions.
MitoPilot only needs a curation database for protein-coding genes. rRNA and tRNA curation does not utilize reference alignment.
Using a custom curation database with MitoPilot
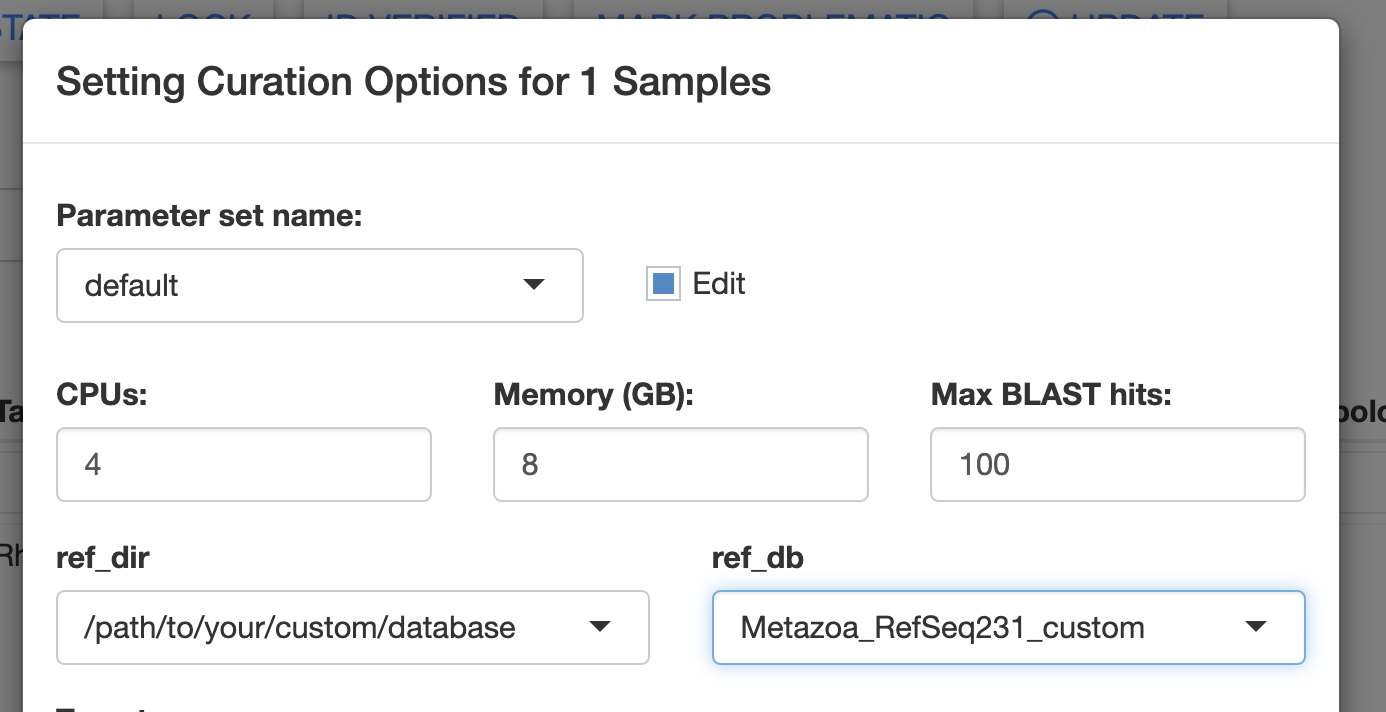
To use a custom curation database, you need to specify the database directory path in the ref_dir field in the Curate Opts. window of the Annotate module. This path will be printed by the MitoPilot::custom_curation_db function.
You should also specify “Metazoa_RefSeq231_custom” or “Chordata_custom” in the ref_db field, depending on which base database you used with the MitoPilot::custom_curation_db function.